DemoKin
DemoKin uses matrix demographic methods to compute
expected (average) kin counts from demographic rates under a range of
scenarios and assumptions. The package is an R-language implementation
of Caswell (2019, 2020, 2022), and Caswell and Song (2021). It draws on
previous theoretical development by Goodman, Keyfitz and Pullum
(1974).
Installation
You can install the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("IvanWilli/DemoKin")
Usage
Consider an average Swedish woman called ‘Focal.’ For this exercise,
we assume a female closed population in which everyone experiences the
Swedish 2015 mortality and fertility rates at each age throughout their
life; i.e., the ‘time-invariant’ assumption in Caswell (2019).
We then ask:
What is the expected number of relatives of Focal over her life
course?
Let’s explore this using the Swedish data already included with
DemoKin.
library(DemoKin)
swe_surv_2015 <- swe_px[,"2015"]
swe_asfr_2015 <- swe_asfr[,"2015"]
swe_2015 <- kin(p = swe_surv_2015, f = swe_asfr_2015, time_invariant = TRUE)
p is the survival probability by age from a life table and
f are the age specific fertility ratios by age (see
?kin for details).
Now, we can visualize the implied kin counts (i.e., the average
number of living kin) of Focal at age 35 using a network or ‘Keyfitz’
kinship diagram with the function plot_diagram:
# We need to reformat the data a little bit
kin_total <- swe_2015$kin_summary
# Keep only data for Focal's age 35
kin_total <- kin_total[kin_total$age_focal == 35 , c("kin", "count_living")]
names(kin_total) <- c("kin", "count")
plot_diagram(kin_total, rounding = 2)
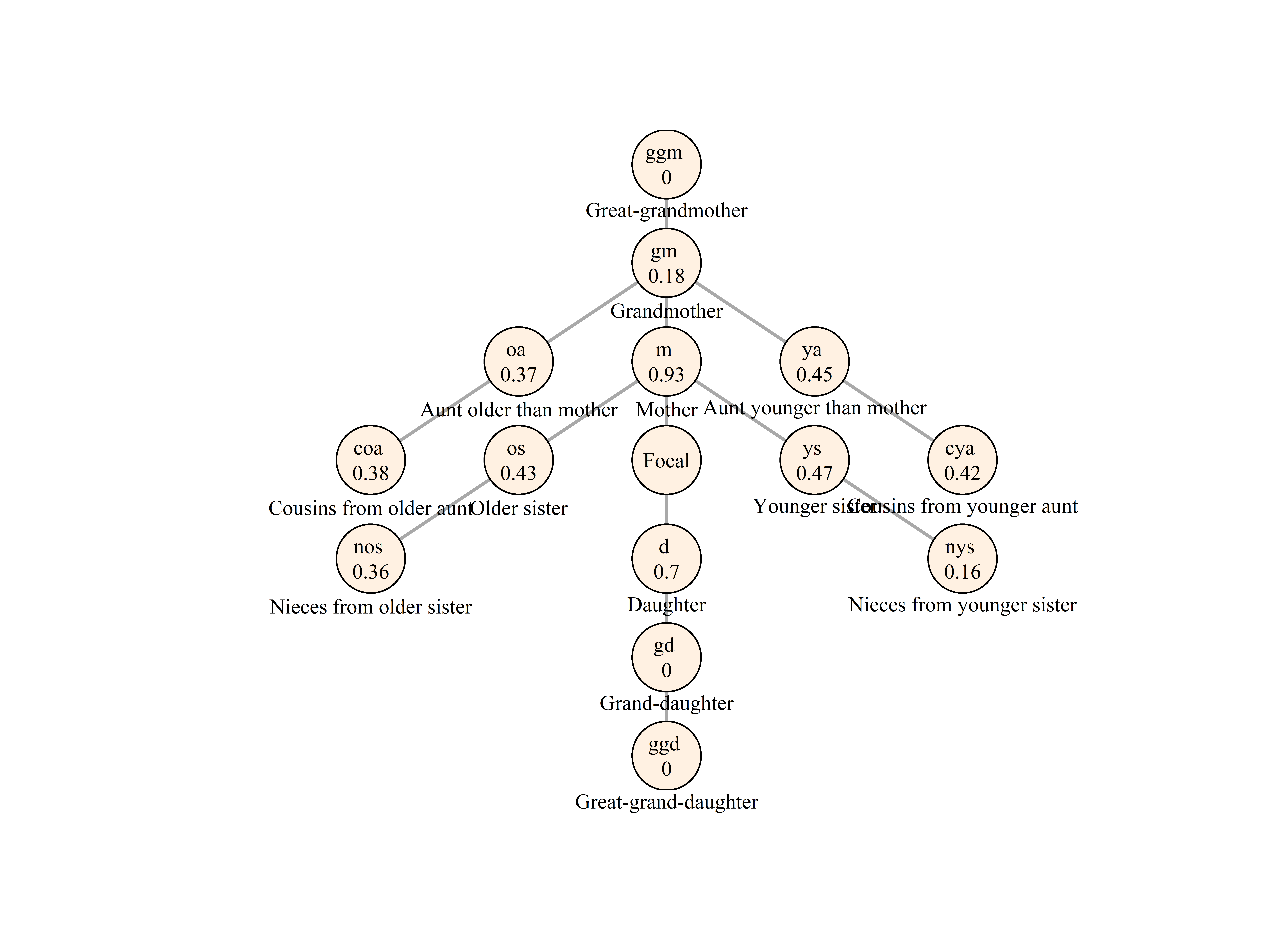
Relatives are identified by a unique code:
| coa |
Cousins from older aunts |
| cya |
Cousins from younger aunts |
| c |
Cousins |
| d |
Daughters |
| gd |
Grand-daughters |
| ggd |
Great-grand-daughters |
| ggm |
Great-grandmothers |
| gm |
Grandmothers |
| m |
Mother |
| nos |
Nieces from older sisters |
| nys |
Nieces from younger sisters |
| n |
Nieces |
| oa |
Aunts older than mother |
| ya |
Aunts younger than mother |
| a |
Aunts |
| os |
Older sisters |
| ys |
Younger sisters |
| s |
Sisters |
Vignette
For more details, including an extension to time-variant rates,
deceased kin, and multi-state models in a one-sex framework, see
vignette("Reference_OneSex", package = "DemoKin"). For
two-sex models, see
vignette("Reference_TwoSex", package = "DemoKin"). If the
vignette does not load, you may need to install the package as
devtools::install_github("IvanWilli/DemoKin", build_vignettes = T).
Citation
Williams, Iván; Alburez-Gutierrez, Diego; Song, Xi; and Hal Caswell.
(2021) DemoKin: An R package to implement demographic matrix kinship
models. URL: https://github.com/IvanWilli/DemoKin.
Acknowledgments
We thank Silvia Leek from the Max Planck Institute for Demographic
Research for designing the DemoKin logo. The logo includes elements that
have been taken or adapted from
this file, originally by Ansunando, CC BY-SA 4.0
via Wikimedia Commons. Sha Jiang provided useful comments for improving
the package.
Get involved!
DemoKin is under constant development. If you’re
interested in contributing, please get in touch, create an issue, or
submit a pull request. We look forward to hearing from you!
References
Caswell, Hal, and Xi Song. 2021. “The Formal Demography of Kinship III:
Kinship Dynamics with Time-Varying Demographic Rates.”
Demographic
Research 45 (August): 517–46.
https://doi.org/10.4054/DemRes.2021.45.16.
Goodman, Leo A, Nathan Keyfitz, and Thomas W. Pullum. 1974. “Family
Formation and the Frequency of Various Kinship Relationships.”
Theoretical Population Biology, 27.
https://doi.org/10.1016/0040-5809(74)90049-5.

