Tidy Verbs for Dealing with Genomic Data Frames
Handle genomic data within data frames just as you would with
GRanges. This packages provides method to deal with
genomics intervals the “tidy-way” which makes it simpler to integrate in
the the general data munging process. The API is inspired by the popular
bedtools and the genome_join() method from the fuzzyjoin package.
install.packages("tidygenomics")
# Or to get the latest development version
devtools::install_github("const-ae/tidygenomics")Joins 2 data frames based on their genomic overlap. Unlike the
genome_join function it updates the boundaries to reflect
the overlap of the regions.
x1 <- data.frame(id = 1:4,
chromosome = c("chr1", "chr1", "chr2", "chr2"),
start = c(100, 200, 300, 400),
end = c(150, 250, 350, 450))
x2 <- data.frame(id = 1:4,
chromosome = c("chr1", "chr2", "chr2", "chr1"),
start = c(140, 210, 400, 300),
end = c(160, 240, 415, 320))
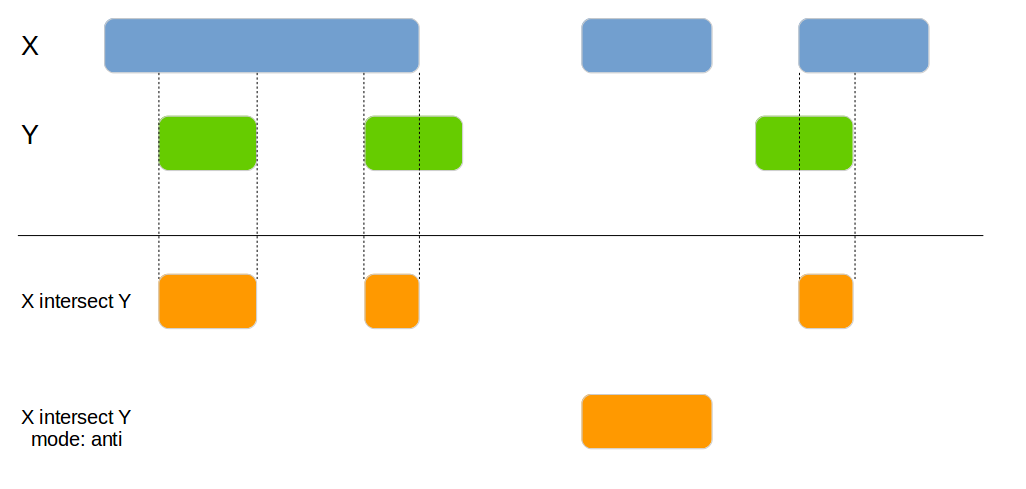
genome_intersect(x1, x2, by=c("chromosome", "start", "end"), mode="both")| id.x | chromosome | id.y | start | end |
|---|---|---|---|---|
| 1 | chr1 | 1 | 140 | 150 |
| 4 | chr2 | 3 | 400 | 415 |
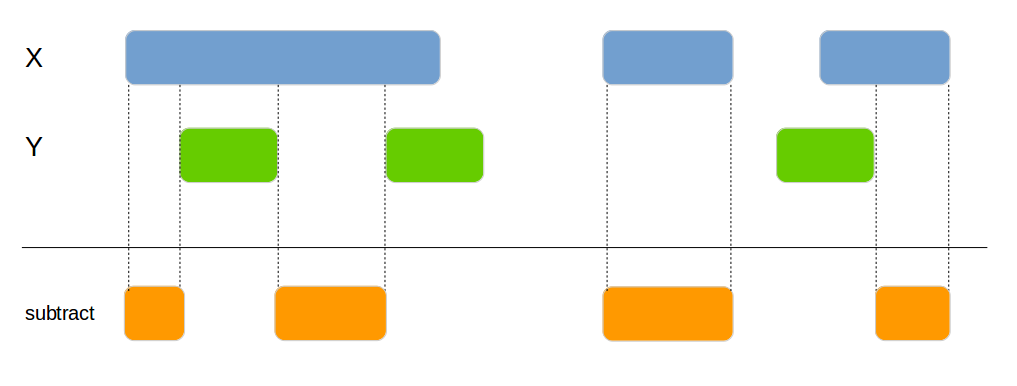
Subtracts one data frame from the other. This can be used to split the x data frame into smaller areas.
x1 <- data.frame(id = 1:4,
chromosome = c("chr1", "chr1", "chr2", "chr1"),
start = c(100, 200, 300, 400),
end = c(150, 250, 350, 450))
x2 <- data.frame(id = 1:4,
chromosome = c("chr1", "chr2", "chr1", "chr1"),
start = c(120, 210, 300, 400),
end = c(125, 240, 320, 415))
genome_subtract(x1, x2, by=c("chromosome", "start", "end"))| id | chromosome | start | end |
|---|---|---|---|
| 1 | chr1 | 100 | 119 |
| 1 | chr1 | 126 | 150 |
| 2 | chr1 | 200 | 250 |
| 3 | chr2 | 300 | 350 |
| 4 | chr1 | 416 | 450 |
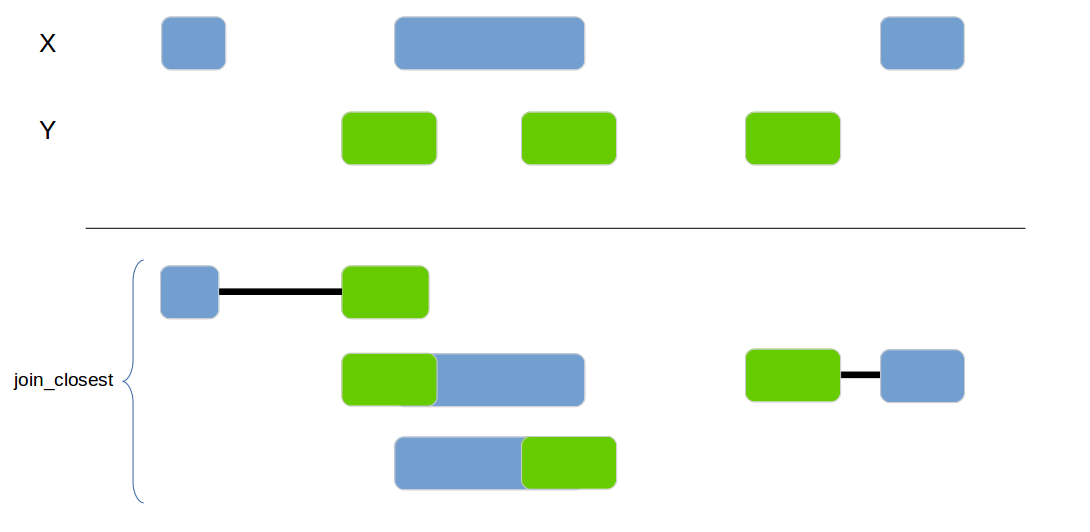
Joins 2 data frames based on their genomic location. If no exact overlap is found the next closest interval is used.
x1 <- data_frame(id = 1:4,
chr = c("chr1", "chr1", "chr2", "chr3"),
start = c(100, 200, 300, 400),
end = c(150, 250, 350, 450))
x2 <- data_frame(id = 1:4,
chr = c("chr1", "chr1", "chr1", "chr2"),
start = c(220, 210, 300, 400),
end = c(225, 240, 320, 415))
genome_join_closest(x1, x2, by=c("chr", "start", "end"), distance_column_name="distance", mode="left")| id.x | chr.x | start.x | end.x | id.y | chr.y | start.y | end.y | distance |
|---|---|---|---|---|---|---|---|---|
| 1 | chr1 | 100 | 150 | 2 | chr1 | 210 | 240 | 59 |
| 2 | chr1 | 200 | 250 | 1 | chr1 | 220 | 225 | 0 |
| 2 | chr1 | 200 | 250 | 2 | chr1 | 210 | 240 | 0 |
| 3 | chr2 | 300 | 350 | 4 | chr2 | 400 | 415 | 49 |
| 4 | chr3 | 400 | 450 | NA | NA | NA | NA | NA |
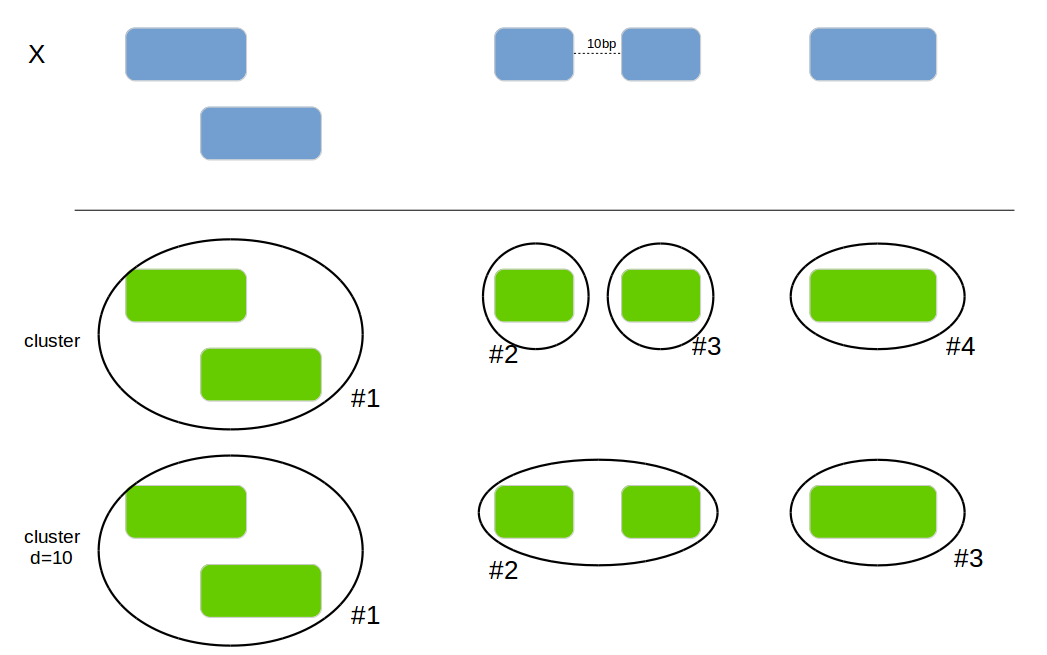
Add a new column with the cluster if 2 intervals are overlapping or
are within the max_distance.
x1 <- data.frame(id = 1:4, bla=letters[1:4],
chromosome = c("chr1", "chr1", "chr2", "chr1"),
start = c(100, 120, 300, 260),
end = c(150, 250, 350, 450))
genome_cluster(x1, by=c("chromosome", "start", "end"))| id | bla | chromosome | start | end | cluster_id |
|---|---|---|---|---|---|
| 1 | a | chr1 | 100 | 150 | 0 |
| 2 | b | chr1 | 120 | 250 | 0 |
| 3 | c | chr2 | 300 | 350 | 2 |
| 4 | d | chr1 | 260 | 450 | 1 |
genome_cluster(x1, by=c("chromosome", "start", "end"), max_distance=10)| id | bla | chromosome | start | end | cluster_id |
|---|---|---|---|---|---|
| 1 | a | chr1 | 100 | 150 | 0 |
| 2 | b | chr1 | 120 | 250 | 0 |
| 3 | c | chr2 | 300 | 350 | 1 |
| 4 | d | chr1 | 260 | 450 | 0 |
Calculates the complement of a genomic region.
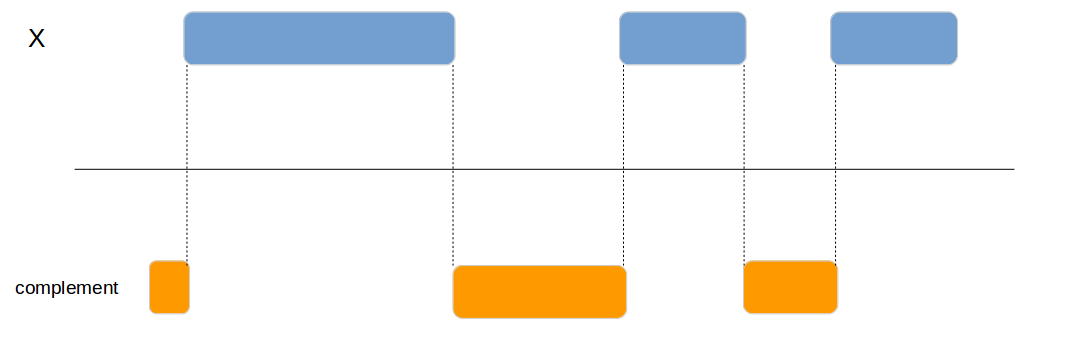
x1 <- data.frame(id = 1:4,
chromosome = c("chr1", "chr1", "chr2", "chr1"),
start = c(100, 200, 300, 400),
end = c(150, 250, 350, 450))
genome_complement(x1, by=c("chromosome", "start", "end"))| chromosome | start | end |
|---|---|---|
| chr1 | 1 | 99 |
| chr1 | 151 | 199 |
| chr1 | 251 | 399 |
| chr2 | 1 | 299 |
Classical join function based on the overlap of the interval. Implemented and maintained in the fuzzyjoin package and documented here only for completeness.

x1 <- data_frame(id = 1:4,
chr = c("chr1", "chr1", "chr2", "chr3"),
start = c(100, 200, 300, 400),
end = c(150, 250, 350, 450))
x2 <- data_frame(id = 1:4,
chr = c("chr1", "chr1", "chr1", "chr2"),
start = c(220, 210, 300, 400),
end = c(225, 240, 320, 415))
fuzzyjoin::genome_join(x1, x2, by=c("chr", "start", "end"), mode="inner")| id.x | chr.x | start.x | end.x | id.y | chr.y | start.y | end.y |
|---|---|---|---|---|---|---|---|
| 2 | chr1 | 200 | 250 | 1 | chr1 | 220 | 225 |
| 2 | chr1 | 200 | 250 | 2 | chr1 | 210 | 240 |
fuzzyjoin::genome_join(x1, x2, by=c("chr", "start", "end"), mode="left")| id.x | chr.x | start.x | end.x | id.y | chr.y | start.y | end.y |
|---|---|---|---|---|---|---|---|
| 1 | chr1 | 100 | 150 | NA | NA | NA | NA |
| 2 | chr1 | 200 | 250 | 1 | chr1 | 220 | 225 |
| 2 | chr1 | 200 | 250 | 2 | chr1 | 210 | 240 |
| 3 | chr2 | 300 | 350 | NA | NA | NA | NA |
| 4 | chr3 | 400 | 450 | NA | NA | NA | NA |
fuzzyjoin::genome_join(x1, x2, by=c("chr", "start", "end"), mode="anti")| id | chr | start | end |
|---|---|---|---|
| 1 | chr1 | 100 | 150 |
| 3 | chr2 | 300 | 350 |
| 4 | chr3 | 400 | 450 |
If you have any additional questions or encounter issues please raise them on the github page.